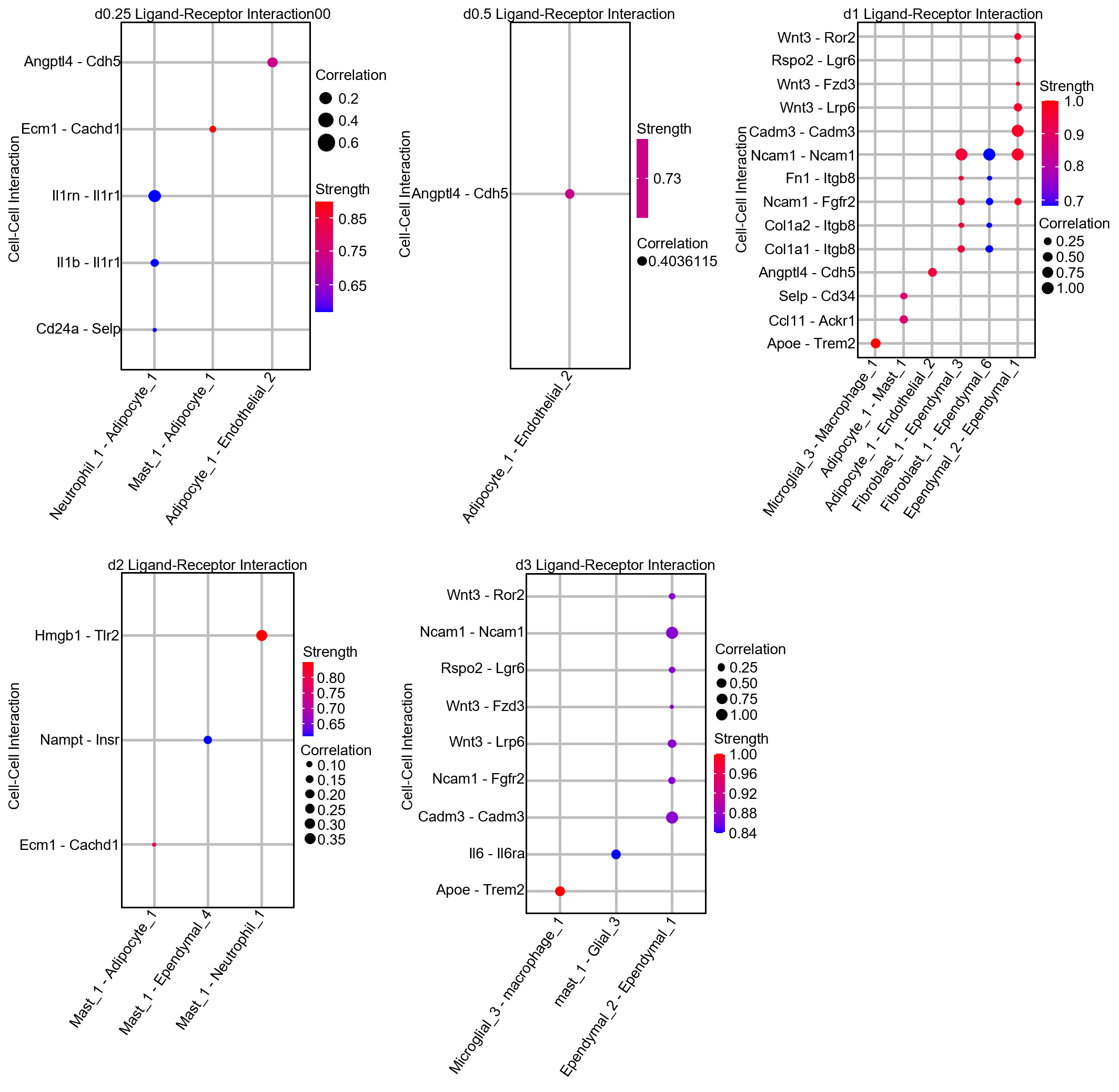DeconvCellLink: Approach for Infer potential deconvolution cell types interactions from mouse bulk RNA-seq data
Version 1.2 (2024-03-17)
Added new functions of receptor-ligand interactions
Version 1.1 (2024-03-15)
Added new functions based on GSEA algorithm
Added multi-threading to optimize running speed
install.packages("remotes")
install.packages("BiocManager")
BiocManager::install("JH-42/DeconvCellLink")
if (!require(BiocManager)) install.packages("BiocManager")
if (!require(remotes)) install.packages("remotes")
if (!require(bcv)) install.packages("bcv")
if (!require(SSMD)) BiocManager::install("JH-42/SSMD")
if (!require(clusterProfiler)) BiocManager::install("clusterProfiler")
if (!require(CBNplot)) BiocManager::install("noriakis/CBNplot")
if (!require(pheatmap)) install.packages("pheatmap")
if (!require(reshape2)) install.packages("reshape2")
if (!require(bnlearn)) install.packages("bnlearn")
if (!require(igraph)) install.packages("igraph")
if (!require(ggraph)) install.packages("ggraph")
if (!require(bnviewer)) install.packages("bnviewer")
if (!require(dplyr)) install.packages("dplyr")
if (!require(tidyr)) install.packages("tidyr")
if (!require(stringr)) install.packages("stringr")
if (!require(nnls)) install.packages("nnls")
if (!require(org.Mm.eg.db)) BiocManager::install("org.Mm.eg.db")
if (!require(parallel)) BiocManager::install("parallel")
if (!require(doParallel)) BiocManager::install("doParallel")
if (!require(foreach)) BiocManager::install("foreach")
#The exp_dat and geneList should use Gene Symbols.
#single tissue cell types
DCL_obj <- DCL_net(exp_dat,geneList,tissueType = "Inflammatory")
#multiple tissue cell types
DCL_obj <- DCL_GSEA_net(expression_data = exp,geneList = geneList,tissueType = NULL,mult = T,
mult_tissue = c("Inflammatory","Central Nervous System"),numCores = 12)
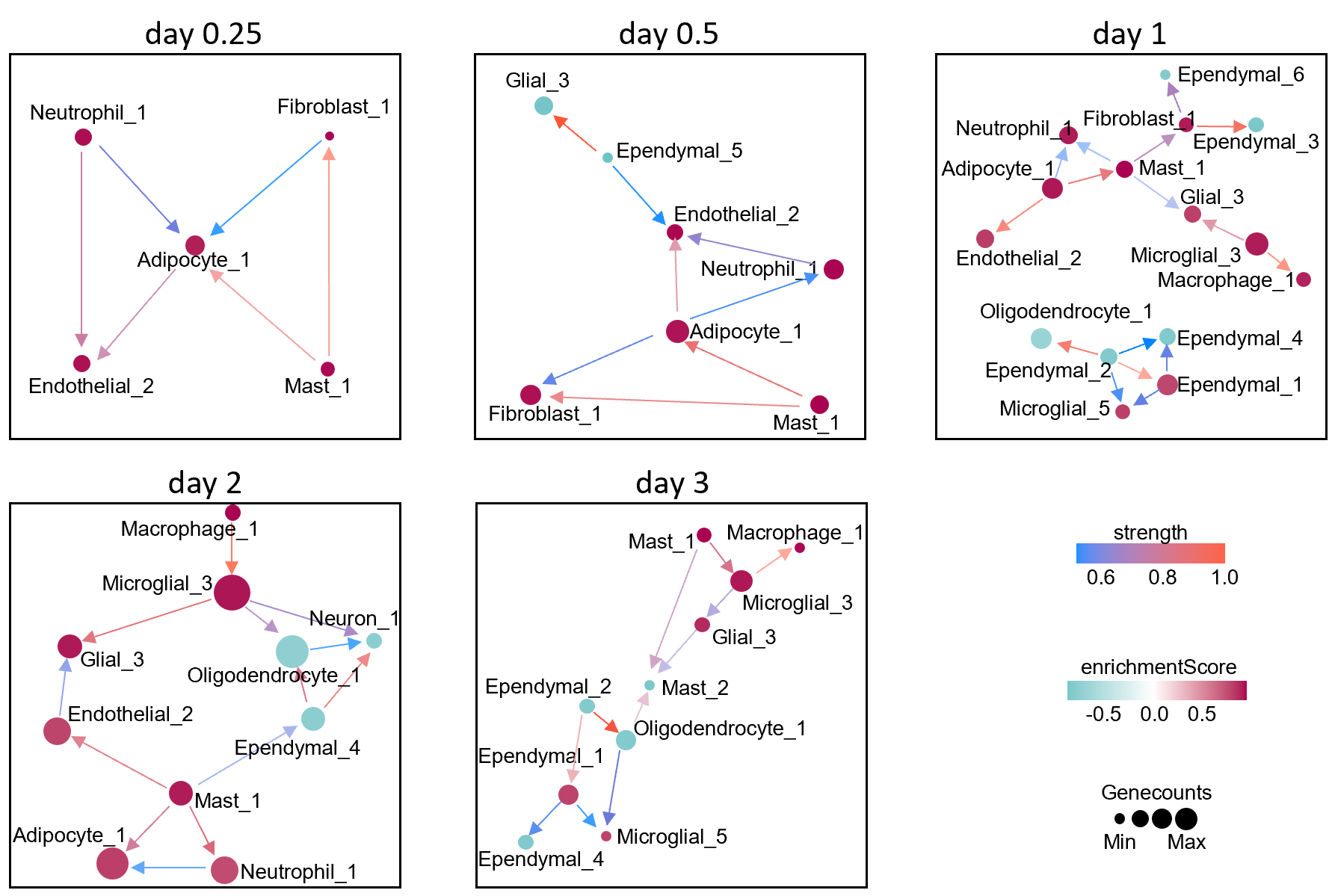
DCL_obj$bnObject
pheatmap::pheatmap(DCL_obj$cells_proportion,scale = "row")
#LR_plot
LR_plot<-DCL_GSEA_net_interaction(DCL_obj,deg = deg, expression_data = exp,cor_method = "pearson",cor_threshold = 0)#deg = deg list
LR_plot$LR_plot
exp_datA data matrix containing the expression data log(CPM/TPM).geneListA gene list, such as a list of differentially expressed genes (DEgenes).tissueTypeThe tissue type of the expression data. Choose one of the following: 'Inflammatory', 'Central Nervous System', 'Hematopoietic System', 'Blood'.multWhether to use multi-tissue annotations.mult_tissueWhich tissue annotations to use.numCoresHow many cores to use.
An object of class is also invisibly returned. This is a list containing the following components:
bnObjectDeconvolution cell link from Bayesian Network.cells_proportionDeconvolution Cell proportion from SSMD.
If you use PathDeconv in your research, please cite it as follows:
Junhao Wang, Zhaoqian Zhong etc.al . (2024). DeconvCellLink: Approach for Infer potential deconvolution cell types interactions from mouse bulk RNA-seq data. GitHub repository: https://github.com/JH-42/DeconvCellLink