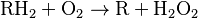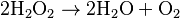Peroxisome
Peroxisomes IPA: [pɛɜˈɹɒksɪˌsoʊmz][1] (also called microbodies) are organelles found in virtually all eukaryotic cells.[2] They are involved in the catabolism of very long chain fatty acids, branched chain fatty acids, D-amino acids, and polyamines, reduction of reactive oxygen species - specifically hydrogen peroxide[3] - and biosynthesis of plasmalogens, i.e. ether phospholipids critical for the normal function of mammalian brains and lungs.[4] They also contain approximately 10% of the total activity of two enzymes in the pentose phosphate pathway, which is important for energy metabolism.[4] It is vigorously debated if peroxisomes are involved in isoprenoid and cholesterol synthesis in animals.[4] Other known peroxisomal functions include the glyoxylate cycle in germinating seeds ("glyoxysomes"), photorespiration in leaves,[5] glycolysis in trypanosomes ("glycosomes"), and methanol and/or amine oxidation and assimilation in some yeasts.
Peroxisomes were identified as organelles by the Belgian cytologist Christian de Duve in 1967[6] after they had been first described by a Swedish doctoral student, J. Rhodin in 1954.[7]
Contents
Metabolic functions
A major function of the peroxisome is the breakdown of very long chain fatty acids through beta-oxidation. In animal cells, the long fatty acids are converted to medium chain fatty acids, which are subsequently shuttled to mitochondria where they are eventually broken down to carbon dioxide and water. In yeast and plant cells, this process is exclusive for the peroxisomes.[8]
The first reactions in the formation of plasmalogen in animal cells also occur in peroxisomes. Plasmalogen is the most abundant phospholipid in myelin. Deficiency of plasmalogens causes profound abnormalities in the myelination of nerve cells, which is one reason why many peroxisomal disorders affect the nervous system.[8] Peroxisomes also play a role in the production of bile acids important for the absorption of fats and fat-soluble vitamins, such as vitamins A and K. Skin disorders are features of genetic disorders affecting peroxisome function as a result.
Peroxisomes contain oxidative enzymes, such as catalase, D-amino acid oxidase, and uric acid oxidase.[9] However the last enzyme is absent in humans, explaining the disease known as gout, caused by the accumulation of uric acid. Certain enzymes within the peroxisome, by using molecular oxygen, remove hydrogen atoms from specific organic substrates (labeled as R), in an oxidative reaction, producing hydrogen peroxide (H2O2, itself toxic):
Catalase, another peroxisomal enzyme, uses this H2O2 to oxidize other substrates, including phenols, formic acid, formaldehyde, and alcohol, by means of the peroxidation reaction:
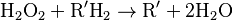 , thus eliminating the poisonous hydrogen peroxide in the process.
, thus eliminating the poisonous hydrogen peroxide in the process.
This reaction is important in liver and kidney cells, where the peroxisomes detoxify various toxic substances that enter the blood. About 25% of the ethanol alcohol humans drink is oxidized to acetaldehyde in this way.[8] In addition, when excess H2O2 accumulates in the cell, catalase converts it to H2O through this reaction:
In higher plants, peroxisomes contain also a complex battery of antioxidative enzymes such as superoxide . dismutase, the components of the ascorbate-glutathione cycle, and the NADP-dehydrogenases of the pentose-phosphate pathway. It has been demonstrated that peroxisomes generate superoxide (O2•−) and nitric oxide (•NO) radicals.[10][11]
The peroxisome of plant cells is polarised when fighting fungal penetration. Infection causes a glucosinolate molecule to play an antifungal role to be made and delivered to the outside of the cell through the action of the peroxisomal proteins (PEN2 and PEN3).[12]
Peroxisome assembly
Peroxisomes can be derived from the endoplasmic reticulum and replicate by fission.[13] Peroxisome matrix proteins are translated in the cytoplasm prior to import. Specific amino acid sequences (PTS or peroxisomal targeting signal) at the C-terminus (PTS1) or N-terminus (PTS2) of peroxisomal matrix proteins signals them to be imported into the organelle. There are at least 32 known peroxisomal proteins, called peroxins,[14] which participate in the process of peroxisome assembly. Proteins do not have to unfold to be imported into the peroxisome. The protein receptors, the peroxins PEX5 and PEX7, accompany their cargoes (containing a PTS1 or a PTS2 amino acid sequence, respectively) all the way into the peroxisome where they release the cargo and then return to the cytosol - a step named recycling. A model describing the import cycle is referred to as the extended shuttle mechanism.[15] There is now evidence that ATP hydrolysis is required for the recycling of receptors to the cytosol. Also, ubiquitination appears to be crucial for the export of PEX5 from the peroxisome, to the cytosol.
Associated medical conditions
Peroxisomal disorders are a class of medical conditions that typically affect the human nervous system as well as many other organ systems. Two common examples are X-linked adrenoleukodystrophy and peroxisome biogenesis disorders.[16][17]
Genes
PEX genes encode the protein machinery ("peroxins") required for proper peroxisome assembly, as described above. Membrane assembly and maintenance requires three of these (peroxins 3, 16, and 19) and may occur without the import of the matrix (lumen) enzymes. Proliferation of the organelle is regulated by Pex11p.
Genes that encode peroxin proteins include: PEX1, PEX2 - PXMP3, PEX3, PEX5, PEX6, PEX7, PEX10, PEX11A, PEX11B, PEX11G, PEX12, PEX13, PEX14, PEX16, PEX19, PEX26, PEX28, PEX30, and PEX31.
Evolutionary origins
The protein content of peroxisomes varies across species, but the presence of proteins common to many species has been used to suggest an endosymbiotic origin; that is, peroxisomes evolved from bacteria that invaded larger cells as parasites, and very gradually evolved a symbiotic relationship.[18] However, this view has been challenged by recent discoveries.[19] For example, peroxisome-less mutants can restore peroxisomes upon introduction of the wild-type gene.
Two independent evolutionary analyses of the peroxisomal proteome found homologies between the peroxisomal import machinery and the ERAD pathway in the endoplasmic reticulum,[20][21] along with a number of metabolic enzymes that were likely recruited from the mitochondria.[21] Recently, it has been suggested that the peroxisome may have had an actinobacterial origin,[22] however, this is controversial.[23]
Other organelles of the microbody family related to peroxisomes include glyoxysomes of plants and filamentous fungi, glycosomes of kinetoplastids[24] and Woronin bodies of filamentous fungi.
References
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 4.0 4.1 4.2 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 8.0 8.1 8.2 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 21.0 21.1 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
Further reading
- Lua error in package.lua at line 80: module 'strict' not found.
- Lua error in package.lua at line 80: module 'strict' not found.
External links
| Wikiversity has learning materials about Peroxisomes at |
| Wikimedia Commons has media related to Peroxisomes. |
![]() This article incorporates public domain material from the NCBI document "Science Primer". This article incorporates text from the public domain Pfam and InterPro IPR006708
This article incorporates public domain material from the NCBI document "Science Primer". This article incorporates text from the public domain Pfam and InterPro IPR006708