Sirtuin
| Sir2 family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
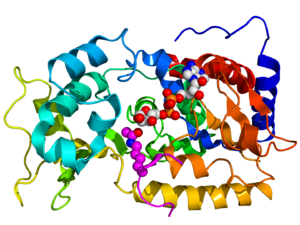
Crystallographic structure of yeast sir2 (rainbow colored cartoon, N-terminus = blue, C-terminus = red) complexed with ADP (space-filling model, carbon = white, oxygen = red, nitrogen = blue, phosphorus = orange) and a histone H4 peptide (magenta) containing an acylated lysine residue (displayed as spheres).[1]
|
|||||||||
| Identifiers | |||||||||
| Symbol | SIR2 | ||||||||
| Pfam | PF02146 | ||||||||
| Pfam clan | CL0085 | ||||||||
| InterPro | IPR003000 | ||||||||
| PROSITE | PS50305 | ||||||||
| SCOP | 1j8f | ||||||||
| SUPERFAMILY | 1j8f | ||||||||
|
|||||||||
Sirtuin or Sir2 proteins are a class of proteins that possess either mono-ADP-ribosyltransferase, or deacylase activity, including deacetylase, desuccinylase, demalonylase, demyristoylase and depalmitoylase activity.[2][3][4][5] Sirtuins regulate important biological pathways in bacteria, archaea and eukaryotes. The name Sir2 comes from the yeast gene 'silent mating-type information regulation 2',[6] the gene responsible for cellular regulation in yeast.
Sirtuins have been implicated in influencing a wide range of cellular processes like aging, transcription, apoptosis, inflammation[7] and stress resistance, as well as energy efficiency and alertness during low-calorie situations.[8] Sirtuins can also control circadian clocks and mitochondrial biogenesis.
Yeast Sir2 and some, but not all, sirtuins are protein deacetylases. Unlike other known protein deacetylases, which simply hydrolyze acetyl-lysine residues, the sirtuin-mediated deacetylation reaction couples lysine deacetylation to NAD hydrolysis. This hydrolysis yields O-acetyl-ADP-ribose, the deacetylated substrate and nicotinamide, itself an inhibitor of sirtuin activity. The dependence of sirtuins on NAD links their enzymatic activity directly to the energy status of the cell via the cellular NAD:NADH ratio, the absolute levels of NAD, NADH or nicotinamide or a combination of these variables.
Contents
Species distribution
Whereas bacteria and archaea encode either one or two sirtuins, eukaryotes encode several sirtuins in their genomes. In yeast, roundworms, and fruitflies, sir2 is the name of the sirtuin-type protein.[9] This research started in 1991 by Leonard Guarente of MIT.[10][11] Mammals possess seven sirtuins (SIRT1-7) that occupy different subcellular compartments such as the nucleus (SIRT1, -2, -6, -7), cytoplasm (SIRT1 and SIRT2) and the mitochondria (SIRT3, -4 and -5).
Types
The first sirtuin was identified in yeast (a lower eukaryote) and named sir2. In more complex mammals, there are seven known enzymes that act in cellular regulation, as sir2 does in yeast. These genes are designated as belonging to different classes, depending on their amino acid sequence structure.[12][13] Several Gram positive prokaryotes as well as the Gram negative hyperthermophilic bacterium Thermotoga maritima possess sirtuins that are intermediate in sequence between classes. These are placed in class U.[12]
| Class | Subclass | Species | Intracellular location |
Activity | Function | |||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Yeast | Mouse | Human | |||||
| I | a | Sir2 or Sir2p, Hst1 or Hst1p |
Sirt1 | SIRT1 | nucleus, cytoplasm | deacetylase | metabolism inflammation |
|
| b | Hst2 or Hst2p | Sirt2 | SIRT2 | cytoplasm | deacetylase | cell cycle, tumorigenesis |
||
| Sirt3 | SIRT3 | nucleus and mitochondria |
deacetylase | metabolism | ||||
| c | Hst3 or Hst3p, Hst4 or Hst4p |
|||||||
| II | Sirt4 | SIRT4 | mitochondria | ADP-ribosyl transferase |
insulin secretion | |||
| III | Sirt5 | SIRT5 | mitochondria | demalonylase, desuccinylase and deacetylase | ammonia detoxification | |||
| IV | a | Sirt6 | SIRT6 | nucleus | Demyristoylase, depalmitoylase, ADP-ribosyl transferase and deacetylase |
DNA repair, metabolism, TNF secretion |
||
| b | Sirt7 | SIRT7 | nucleolus | deacetylase | rDNA transcription |
|||
| U | cobB[14] | regulation of acetyl-CoA synthetase[15] |
metabolism | |||||
Sirtuin list based on North/Verdin diagram.[16]
Clinical significance
Sirtuin activity is inhibited by nicotinamide, which binds to a specific receptor site,[17] so it is thought that drugs that interfere with this binding should increase sirtuin activity. Development of new agents that would specifically block the nicotinamide-binding site could provide an avenue for development of newer agents to treat degenerative diseases such as cancer, diabetes, atherosclerosis, and gout.[18][19]
Diabetes
Sirtuins have been proposed as a chemotherapeutic target for type II diabetes mellitus.[20]
Aging
Preliminary studies with resveratrol, a possible SIRT1 activator, have led some scientists to speculate that resveratrol may extend lifespan.[21] Further experiments conducted by Rafael de Cabo et al. showed that resveratrol-mimicking drugs such as SRT1720 could extend the lifespan of obese mice by 44%.[22] Comparable molecules are now undergoing clinical trials in humans.
Cell culture research into the behaviour of the human sirtuin SIRT1 shows that it behaves like the yeast sirtuin Sir2: SIRT2 assists in the repair of DNA and regulates genes that undergo altered expression with age.[23] Adding resveratrol to the diet of mice inhibit gene expression profiles associated with muscle aging and age-related cardiac dysfunction.[24]
A study performed on transgenic mice overexpressing SIRT6, showed an increased lifespan of about 15% in males. The transgenic males displayed lower serum levels of insulin-like growth factor 1 (IGF1) and changes in its metabolism, which may have contributed to the increased lifespan.[25]
See also
- Ageing
- Sir2
- Resveratrol
- Biological immortality
- Caloric restriction
- Trichostatin A
- Histone deacetylases or HDACs
References
- ↑ PDB: 1szd; Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ EntrezGene 23410
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 12.0 12.1 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
External links
- Sirtuins at the US National Library of Medicine Medical Subject Headings (MeSH)
- Lua error in package.lua at line 80: module 'strict' not found.