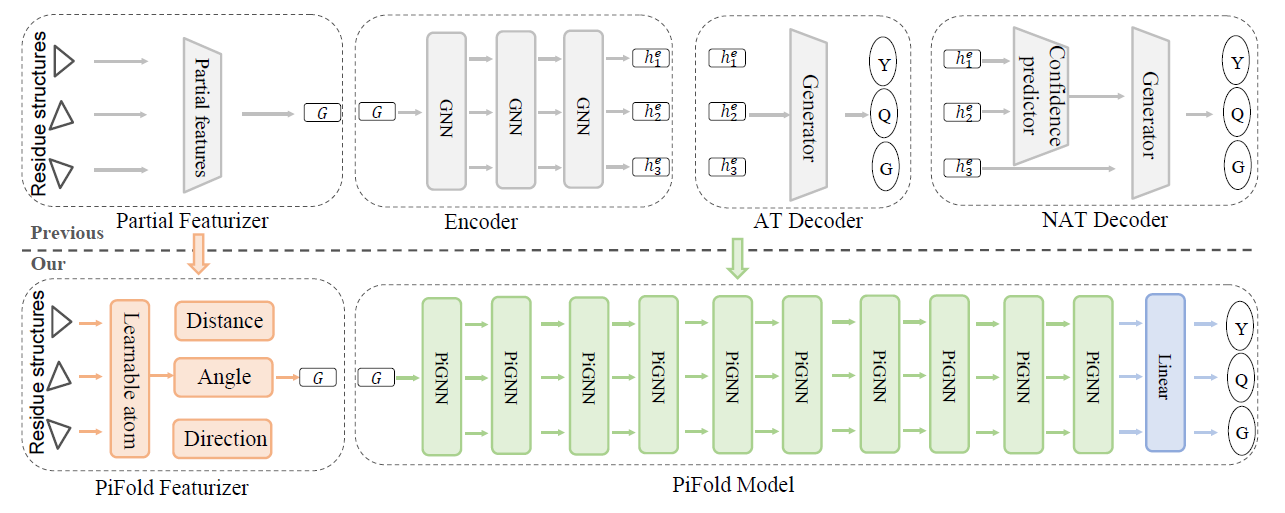
An unofficial re-implementation of PiFold, a fast inverse-folding algorithm for protein sequence design, in PyTorch.
$ pip install pifold-pytorchfrom pifold_pytorch import PiFold
model = PiFold(
d_node=165, d_edge=525, d_emb=128, d_rbf=16,
n_heads=4, num_layers=10, n_virtual_atoms=3, n_neighbors=30
)
node = torch.randn(100, 165) # Node features
edge = torch.randn(3000, 525) # Edge features
edge_index = torch.randint(0, 100, (2, 3000)) # Edge indices
batch_idx = torch.zeros(100, dtype=torch.long) # Batch indices
output = model(node, edge, edge_index, batch_idx)
output.shape # (100, 20), Probabilities for amino acids at each position.Logs for train/validation of PiFold with CATH 4.2 dataset can be found here. Early stopping with patience of 7 epochs was used.
| Model | Perplexity (test) | Per-protein median recovery (test) |
|---|---|---|
| Paper (10 layers) | 4.55 | 51.66 |
| Reproduction (24 layers, N(0, 1/25) noise at dist) | 4.611 | 52.52 |
| Reproduction (16 layers) | 4.702 | 52.27 |
| Reproduction (10 layers) | 4.645 | 51.28 |
| Reproduction (10 layers, N(0, 1/25) noise at dist) | 4.656 | 51.59 |
| Reproduction (16 layers, N(0, 1/25) noise at dist) | 4.666 | 51.68 |
@article{gao2022pifold,
title={PiFold: Toward effective and efficient protein inverse folding},
author={Gao, Zhangyang and Tan, Cheng and Li, Stan Z},
journal={arXiv preprint arXiv:2209.12643},
year={2022}
}