Ammonia transporter
| Ammonia transporter | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|||||||||
| Identifiers | |||||||||
| Symbol | AmtB | ||||||||
| Pfam | PF00909 | ||||||||
| InterPro | IPR001905 | ||||||||
| TCDB | 1.A.11 | ||||||||
| OPM superfamily | 13 | ||||||||
| OPM protein | 2ns1 | ||||||||
|
|||||||||
Ammonia transporters are structurally related membrane transport proteins called Amt proteins (ammonia transporters) in bacteria and plants, methylammonium/ammonium permeases (MEPs) in yeast, or Rhesus proteins (Rh) in chordates. In humans, the RhAG, RhBG, and RhCG Rhesus proteins constitute solute carrier family 42[2] whilst RhD and RhCE form the Rh blood group system. The three-dimensional structure of the ammonia transport protein AmtB from Escherichia coli has been determined by x-ray crystallography[3][4] revealing a hydrophobic ammonia channel.[5] The human RhCG ammonia transporter was found to have a similar ammonia-conducting channel structure.[1] It was proposed[citation needed] that the erythrocyte Rh complex is a heterotrimer of RhAG, RhD, and RhCE subunits in which RhD and RhCE might play roles in anchoring the ammonia-conducting RhAG subunit to the cytoskeleton. Based on reconstitution experiments, purified RhCG subunits alone can function to transport ammonia.[6] RhCG is required for normal acid excretion by the mouse kidney[7] and epididymis.[8]
Structure
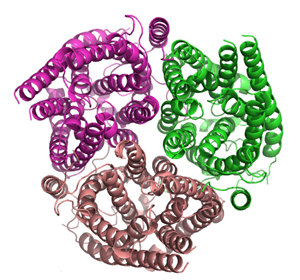
The structure of the ammonia channel from E. coli,[3][4] was, at the time of its publication, the highest resolution structure of any integral membrane protein. It shows a trimer of subunits, each made up of 11 transmembrane helices and containing a pseudo two-fold symmetry.[9] Each monomer contains a hydrophobic ammonia conducting channel. Within the channel are two signature histidine residues which are entirely conserved in the family of active transporters.
While prokaryotic ammonia channel proteins have an N-terminal region which acts as a signal sequence and is cleaved in the mature protein,[10] the Rhesus glycoproteins retain this as a 12th transmembrane helix in the mature protein.[1]
Regulation
In E. coli the AmtB gene is expressed only under limiting nitrogen levels to yield the AmtB protein. It is co-expressed with the GlnK gene which encodes a PII protein. This protein is trimeric also and remains in the cytoplasm.[11] It is covalently modified by a uridyl group. When nitrogen levels outside the cell rise, the ammonia channel must be deactivated to prevent excessive ammonia entering the cell (where ammonia would be combined with glutamate to make glutamine, utilising ATP and thereby depleting the cell's ATP reserves). This deactivation is achieved by de-uridylation of the GlnK protein which then binds to the cytoplasmic face of AmtB and inserts a loop into the ammonia conducting pore. At the tip of this loop is an arginine residue which sterically blocks the channel.[12]
References
- ↑ 1.0 1.1 1.2 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 3.0 3.1 1xqe; Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 4.0 4.1 2u7c; Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 2nuu; Lua error in package.lua at line 80: module 'strict' not found.