Molecular clock
Lua error in package.lua at line 80: module 'strict' not found. The molecular clock is a technique that uses the mutation rate of biomolecules to deduce the time in prehistory when two or more life forms diverged. The biomolecular data used for such calculations are usually nucleotide sequences for DNA or amino acid sequences for proteins. The benchmarks for determining the mutation rate are often fossil or archaeological dates. The molecular clock was first tested in 1962 on the haemoglobin protein variants of various animals, and is commonly used in molecular evolution to estimate times of speciation or radiation. It is sometimes called a gene clock or an evolutionary clock.
Contents
Early discovery and genetic equidistance
The notion of the existence of a so-called "molecular clock" was first attributed to Emile Zuckerkandl and Linus Pauling who, in 1962, noticed that the number of amino acid differences in hemoglobin between different lineages changes roughly linearly with time, as estimated from fossil evidence.[1] They generalized this observation to assert that the rate of evolutionary change of any specified protein was approximately constant over time and over different lineages (based on the molecular clock hypothesis (MCH)).
The genetic equidistance phenomenon was first noted in 1963 by Emanuel Margoliash, who wrote: "It appears that the number of residue differences between cytochrome c of any two species is mostly conditioned by the time elapsed since the lines of evolution leading to these two species originally diverged. If this is correct, the cytochrome c of all mammals should be equally different from the cytochrome c of all birds. Since fish diverges from the main stem of vertebrate evolution earlier than either birds or mammals, the cytochrome c of both mammals and birds should be equally different from the cytochrome c of fish. Similarly, all vertebrate cytochrome c should be equally different from the yeast protein."[2] For example, the difference between the cytochrome c of a carp and a frog, turtle, chicken, rabbit, and horse is a very constant 13% to 14%. Similarly, the difference between the cytochrome c of a bacterium and yeast, wheat, moth, tuna, pigeon, and horse ranges from 64% to 69%. Together with the work of Emile Zuckerkandl and Linus Pauling, the genetic equidistance result directly led to the formal postulation of the molecular clock hypothesis in the early 1960s.[3] Genetic equidistance has often been used to infer equal time of separation of different sister species from an outgroup.[4][5]
Later Allan Wilson and Vincent Sarich built upon this work.
Relationship with neutral theory
The observation of a clock-like rate of molecular change was originally purely phenomenological. Later, the work of Motoo Kimura[6] developed the neutral theory of molecular evolution, which predicted a molecular clock. Let there be N individuals, and to keep this calculation simple, let the individuals be haploid (i.e. have one copy of each gene). Let the rate of neutral mutations (i.e. mutations with no effect on fitness) in a new individual be 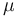 . The probability that this new mutation will become fixed in the population is then 1/N, since each copy of the gene is as good as any other. Every generation, each individual can have new mutations, so there are
. The probability that this new mutation will become fixed in the population is then 1/N, since each copy of the gene is as good as any other. Every generation, each individual can have new mutations, so there are 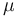 N new neutral mutations in the population as a whole. That means that each generation,
N new neutral mutations in the population as a whole. That means that each generation, 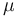 new neutral mutations will become fixed. If most changes seen during molecular evolution are neutral, then fixations in a population will accumulate at a clock-rate that is equal to the rate of neutral mutations in an individual.
new neutral mutations will become fixed. If most changes seen during molecular evolution are neutral, then fixations in a population will accumulate at a clock-rate that is equal to the rate of neutral mutations in an individual.
Calibration
The molecular clock alone can only say that one time period is twice as long as another: it cannot assign concrete dates. To achieve this, the molecular clock must first be calibrated against independent evidence about dates, such as the fossil record.[7] For viral phylogenetics and ancient DNA studies – two areas of evolutionary biology where it is possible to sample sequences over an evolutionary timescale – the dates of the intermediate samples can be used to more precisely calibrate the molecular clock.
Non-constant rate of molecular clock
Sometimes only a single divergence date can be estimated from fossils, with all other dates inferred from that. Other sets of species have abundant fossils available, allowing the MCH of constant divergence rates to be tested. DNA sequences experiencing low levels of negative selection showed divergence rates of 0.7–0.8% per Myr in bacteria, mammals, invertebrates, and plants.[8] In the same study, genomic regions experiencing very high negative or purifying selection (encoding rRNA) were considerably slower (1% per 50 Myr).
In addition to such variation in rate with genomic position, since the early 1990s, variation among taxa has proven fertile ground for research too,[9] even over comparatively short periods of evolutionary time (for example mockingbirds[10]). Tube-nosed seabirds have molecular clocks that on average run at half speed of many other birds,[11] possibly due to long generation times, and many turtles have a molecular clock running at one-eighth the speed it does in small mammals or even slower.[12] Effects of small population size are also likely to confound molecular clock analyses. Researchers such as Francisco Ayala have more fundamentally challenged the molecular clock hypothesis.[13][14] According to Ayala's 1999 study, five factors combine to limit the application of molecular clock models:
- Changing generation times (If the rate of new mutations depends at least partly on the number of generations rather than the number of years)
- Population size (Genetic drift is stronger in small populations, and so more mutations are effectively neutral)
- Species-specific differences (due to differing metabolism, ecology, evolutionary history,...)
- Change in function of the protein studied (can be avoided in closely related species by utilizing non-coding DNA sequences or emphasizing silent mutations)
- Changes in the intensity of natural selection.
Molecular clock users have developed workaround solutions using a number of statistical approaches including maximum likelihood techniques and later Bayesian modeling. In particular, models that take into account rate variation across lineages have been proposed in order to obtain better estimates of divergence times. These models are called relaxed molecular clocks[15] because they represent an intermediate position between the 'strict' molecular clock hypothesis and Joseph Felsenstein's many-rates model[16] and are made possible through MCMC techniques that explore a weighted range of tree topologies and simultaneously estimate parameters of the chosen substitution model. It must be remembered that divergence dates inferred using a molecular clock are based on statistical inference and not on direct evidence.
The molecular clock runs into particular challenges at very short and very long timescales. At long timescales, the problem is saturation. When enough time has passed, many sites have undergone more than one change, but it is impossible to detect more than one. This means that the observed number of changes is no longer linear with time, but instead flattens out.
At very short time scales, many differences between samples do not represent fixation of different sequences in the different populations. Instead, they represent alternative alleles that were both present as part of a polymorphism in the common ancestor. The inclusion of differences that have not yet become fixed leads to a potentially dramatic inflation of the apparent rate of the molecular clock at very short timescales.[17][18]
Methods
Bayesian methods can provide more appropriate estimates of divergence times, especially if large datasets—such as those yielded by phylogenomics—are employed.[19]
Uses
The molecular clock technique is an important tool in molecular systematics, the use of molecular genetics information to determine the correct scientific classification of organisms or to study variation in selective forces.
Knowledge of approximately constant rate of molecular evolution in particular sets of lineages also facilitates establishing the dates of phylogenetic events, including those not documented by fossils, such as the divergence of living taxa and the formation of the phylogenetic tree. In these cases—especially over long stretches of time—the limitations of MCH (above) must be considered; such estimates may be off by 50% or more.
See also
- Charles Darwin
- Gene orders
- Human mitochondrial molecular clock
- Mitochondrial Eve and Y-chromosomal Adam
- Models of DNA evolution
- Molecular evolution
- Neutral theory of molecular evolution
References
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Huang, S. (2008) The genetic equidistance result of molecular evolution is independent of mutation rates. J. Comp. Sci. Syst. Biol., 1: 92-102. http://omicsonline.com/ArchiveJCSB/Ab01/JCSB1.092.html
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.

- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
Further reading
- Lua error in package.lua at line 80: module 'strict' not found.
- Lua error in package.lua at line 80: module 'strict' not found.
- Lua error in package.lua at line 80: module 'strict' not found.