Photolyase
| FAD binding domain of DNA photolyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| File:Photolyase 1qnf.png
A deazaflavin photolyase from Anacystis nidulans, illustrating the two light-harvesting cofactors: FADH− (yellow) and 8-HDF (cyan).
|
|||||||||
| Identifiers | |||||||||
| Symbol | FAD_binding_7 | ||||||||
| Pfam | PF03441 | ||||||||
| InterPro | IPR005101 | ||||||||
| PROSITE | PDOC00331 | ||||||||
| SCOP | 1qnf | ||||||||
| SUPERFAMILY | 1qnf | ||||||||
|
|||||||||
Photolyases (EC 4.1.99.3) are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. This enzyme mechanism[1] requires visible light, preferentially from the violet/blue end of the spectrum, and is known as photoreactivation.
Photolyase is a phylogenetically old enzyme which is present and functional in many species, from the bacteria to the fungi to plants and to the animals.[2] Photolyase is particularly important in repairing UV induced damage in plants. The photolyase mechanism is no longer working in humans and other placental mammals who instead rely on the less efficient nucleotide excision repair mechanism.[3]
Photolyases bind complementary DNA strands and break certain types of pyrimidine dimers that arise when a pair of thymine or cytosine bases on the same strand of DNA become covalently linked. These dimers result in a 'bulge' of the DNA structure, referred to as a lesion. The more common covalent linkage involves the formation of a cyclobutane bridge. Photolyases have a high affinity for these lesions and reversibly bind and convert them back to the original bases.
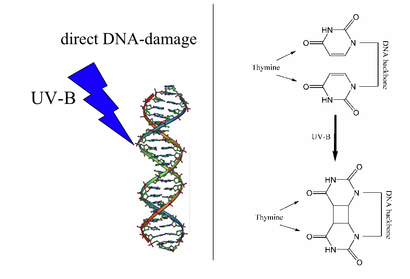
Photolyases are flavoproteins and contain two light-harvesting cofactors. All photolyases contain the two-electron-reduced FADH−; they are divided into two main classes based on the second cofactor, which may be either the pterin methenyltetrahydrofolate (MTHF) in folate photolyases or the deazaflavin 8-hydroxy-7,8-didemethyl-5-deazariboflavin (8-HDF) in deazaflavin photolyases. Although only FAD is required for catalytic activity, the second cofactor significantly accelerates reaction rate in low-light conditions. The enzyme acts by electron transfer in which the reduced flavin FADH− is activated by light energy and acts as an electron donor to break the pyrimidine dimer.[4]
On the basis of sequence similarities DNA photolyases can be grouped into two classes. The first class contains enzymes from Gram-negative and Gram-positive bacteria, the halophilic archaebacteria Halobacterium halobium, fungi and plants. Proteins containing this domain also include Arabidopsis thaliana cryptochromes 1 and 2, which are blue light photoreceptors that mediate blue light-induced gene expression and modulation of circadian rhythms.
The branch named Cryptochrome-Drosophila, Arabidopsis, Synechocystis, Human (Cry-DASH) were previously assumed to have no DNA repair activity because of negligible activity on double-stranded DNA. The study published by A. Sancar and P. Selby provided evidence to suggest this branch of Cryptochromes have photolyase activity with a high degree of specificity for cyclobutane pyrimidine dimers in single-stranded DNA. Their study showed that VcCry1 from Vibrio cholerae, X1Cry from Xenopus laevis, and AtCry3 from Arabidopsis thalina all had photolyase activity on UV irradiated ssDNA in vitro.[2]
Some sunscreens include photolyase in their ingredients, claiming a reparative action on UV-damaged skin.[5]
Human proteins containing this domain
References
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 2.0 2.1 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.