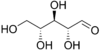
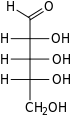
Ribose
|
|
|||
|
|
|||
| Names | |||
|---|---|---|---|
| IUPAC name
(2S,3R,4S,5R)-5-(hydroxymethyl)oxolane-2,3,4-triol
|
|||
| Other names
D-Ribose
|
|||
| Identifiers | |||
| 50-69-1 |
|||
| ChEMBL | ChEMBL1159662 |
||
| ChemSpider | 4470639 aldehydo form D-(−)-Ribose |
||
| DrugBank | DB01936 |
||
| EC Number | 200-059-4 | ||
| Jmol 3D model | aldehydo form D-(−)-Ribose: Interactive image | ||
| PubChem | 5779 5311110 aldehydo form D-(−)-Ribose |
||
|
|||
|
|||
| Properties[1][2] | |||
| C5H10O5 | |||
| Molar mass | 150.13 g/mol | ||
| Appearance | white solid | ||
| Melting point | 95 °C (203 °F; 368 K) | ||
| very soluble[quantify] | |||
|
Chiral rotation ([α]D)
|
−21.5° (H2O) | ||
| Vapor pressure | {{{value}}} | ||
| Related compounds | |||
|
Related aldopentoses
|
Arabinose Xylose Lyxose |
||
|
Related compounds
|
Deoxyribose | ||
|
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
|
|||
| Infobox references | |||
Ribose is an organic compound with the formula C5H10O5; specifically, a pentose monosaccharide (simple sugar) with linear form H−(C=O)−(CHOH)4−H, which has all the hydroxyl groups on the same side in the Fischer projection.
The term may refer to either of two enantiomers. The term usually indicates D-ribose, which occurs widely in nature and is discussed here. Its synthetic mirror image, L-ribose, is not found in nature.
D-Ribose was first reported in 1891 by Emil Fischer. It is a C'-2 carbon epimer of the sugar D-arabinose (both isomers of which are named for their source, gum arabic) and ribose itself is named as a transposition of the name of arabinose.[3]
The ribose β-D-ribofuranose forms part of the backbone of RNA. It is related to deoxyribose, which is found in DNA. Phosphorylated derivatives of ribose such as ATP and NADH play central roles in metabolism. cAMP and cGMP, formed from ATP and GTP, serve as secondary messengers in some signalling pathways.
Structure
Ribose is an aldopentose (a monosaccharide containing five carbon atoms) that, in its open chain form, has an aldehyde functional group at one end. In the conventional numbering scheme for monosaccharides, the carbon atoms are numbered from C1' (in the aldehyde group) to C5'. The deoxyribose derivative found in DNA differs from ribose by having a hydrogen atom in place of the hydroxyl group at C2'. This hydroxyl group performs a function in RNA splicing.
Like many monosaccharides, ribose exists in an equilibrium among 5 forms—the linear form H−(C=O)−(CHOH)4–H and either of the two ring forms: alpha- or beta-ribofuranose (“C3'-endo”), with a five-membered ring, and alpha- or beta-ribopyranose (“C2'-endo”), with a six-membered ring. The beta-ribopyranose form predominates in aqueous solution.[4]
The “D-” in the name D-ribose refers to the stereochemistry of the chiral carbon atom farthest away from the aldehyde group (C4'). In D-ribose, as in all D-sugars, this carbon atom has the same configuration as in D-glyceraldehyde.
Relative abundance of different forms of ribose in solution: β-D-ribopyranose (59%), α-D-ribopyranose (20%), β-D-ribofuranose (13%), α-D-ribofuranose (7%) and open chain (0.1%).[5]
Phosphorylation
In biology, D-ribose must be phosphorylated by the cell before it can be used. Ribokinase catalyzes this reaction by converting D-ribose to D-ribose 5-phosphate. Once converted, D-ribose-5-phosphate is available for the manufacturing of the amino acids tryptophan and histidine, or for use in the pentose phosphate pathway. The absorption of D-ribose is 88–100% in the small intestines (up to 200 mg/kg/h).[6]
References
- ↑ Lua error in package.lua at line 80: module 'strict' not found., 8205
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.