Methylmalonyl-CoA mutase deficiency
| Methylmalonyl-CoA mutase deficiency | |
|---|---|
| Classification and external resources | |
| Specialty | Lua error in Module:Wikidata at line 446: attempt to index field 'wikibase' (a nil value). |
| OMIM | 251000 |
| DiseasesDB | 29509 |
| Patient UK | Methylmalonyl-CoA mutase deficiency |
| Methylmalonyl-CoA mutase | |
|---|---|
| File:Protein MUT PDB 2XIJ.png | |
| Identifiers | |
| Symbol | MUT |
| Alt. symbols | MCM |
| Entrez | 4594 |
| HUGO | 4594 |
| OMIM | 609058 |
| RefSeq | NP_000246 |
| UniProt | P22033 |
| Other data | |
| EC number | 5.4.99.2 |
| Locus | Chr. 6 p21 |
| methylmalonyl-CoA mutase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 5.4.99.2 | ||||||||
| CAS number | Template:CAS | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
|
|||||||||
Methylmalonyl-CoA mutase is a mitochondrial homodimer apoenzyme (EC. 5. 4.99.2) that focuses on the catalysis of malonyl CoA to succinyl CoA. The enzyme is bound to adenosylcobalamin, a hormonal derivative of vitamin B12 in order to function. Methylmalonyl-CoA mutase deficiency[1] is caused by genetic defect in the MUT [2] gene responsible for encoding the enzyme. Deficiency in this enzyme accounts for 60% of the cases of methylmalonic acidemia.[3]
Contents
Symptoms
People with methylmalonyl CoA mutase deficiency exhibit many symptoms similar to other diseases involving inborn errors of metabolism. Sometimes the symptoms appear shortly after birth, but other times the onset of symptoms is later.
Newborn babies experience with vomiting, acidosis, hyperammonemia, hepatomegaly (enlarged livers), hyperglycinemia (high glycine levels), and hypoglycemia (low blood sugar). Later, cases of thrombocytopenia and neutropenia can occur.
In some cases intellectual and developmental disabilities, such as autism, were noted with increased frequency in populations with methylmalonyl-CoA mutase deficiency.[4]
Causes
While methylmalonic acidemia has a variety of causes, both genetic and dietary, methylmalonyl CoA mutase deficiency is an autosomal recessive genetic disorder. Patients suffering from the deficiency either have a complete gene lesion, designated as mut0 or a partial mutation in the form of a frameshift designated as mut-. This frameshift affects the folding of the enzyme rendering its binding domain less effective.[5] Patients with a complete deletion suffer an inactivation of methylmalonyl CoA mutase and exhibit the most sever symptoms of the deficiency, while patients with a partial mutations have a wide range of symptoms. Over 49 different mutations[6] have been discovered for the MUT gene, yet only two appear in any discernible frequency.
Enzymatic activity
Methylmalonyl-CoA mutase catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA, and uses a B12 derived prosthetic group, adenosylcobalamin, in order to accomplish this transfer. The enzyme is a homodimer, located in the mitochondrial matrix. The enzyme is 750 amino acids long, with the a metal ligand binding region to bind to the Cobalt region of adenosylcobalamin.[7]
The enzyme works by cleaving the adenosylcobalamin C-Co(III) bond, giving the carbon and cobalt (III) atoms each an electron. The cobalt then fluctuates between its two oxidation states: Co(II) and Co(III). In this way the adenosylcobalamin works as a reversible free radical generator. The cobalt therefore donates an electron back to the methylmalonyl-CoA backbone in order to transfer the coenzyme A group.[7]
Metabolic activity
Methylmalonyl-CoA mutase is essential to the degradation pathways of many various macromolecules including proteins, odd chain fatty acids, and cholesterol. Methylmalonyl-CoA mutase links the propionyl-CoA degradation byproduct of these macromolecules to the tricarboxylic acid cycle.[7]
For amino acid metabolism, methylmalonyl-CoA mutase works in the degradation pathways of isoleucine, threonine, valine, and methionine. These amino acids are degraded into propanoyl-CoA which is then further degraded into (S)-methylmalonyl-CoA. This substrate must be further metabolized by a very similar enzyme, methylmalonyl-CoA epimerase, which converts the (S) form of methylmalonyl-CoA into the (R) form. This is finally transformed using methylmalonyl-CoA mutase. L-methionine is also metabolized through a longer superpathway (see Figure 2). After transformation to L-homocystein, it is combined with L-serine to make L-cystathione, which is hydrolyzed by cystathione gamma lyase to create 2-oxobutanoate. This substrate is transformed to propionyl-CoA and undergoes the same metabolism previously described for propionyl-CoA.[8]
The cholesterol superpathway follows the degradation of cholesterol down to various substrates, however only a couple of these biotransformed molecules see propionyl-CoA as a byproduct. The conversion of 3,24-dioxocholest-4-en-26-oyl-CoA to 2-oxochol-4-en-24-oyl-CoA sees the release of a propionyl-CoA molecule. Additionally, the conversion of 3-oxo-23,24-bisnorchol-4-en-17-ol-22-oyl-CoA to androst-4-ene-3,17-dione release of a propionyl-CoA molecule. Finally, the degradation of (S)-4-hydroxy-2-oxohexanoate to pyruvate and propanal, in turn releases a propionyl-CoA substrate after the propanal is converted. All these propionyl-CoA substrates are converted to succinyl-CoA following the methylmalonyl pathway For amino acid metabolism, methylmalonyl-CoA mutase works in the degradation pathways of isoleucine, threonine, valine, and methionine. These amino acids are degraded into propanoyl-CoA which is then further degraded into (S)-methylmalonyl-CoA. This substrate must be further metabolized by a very similar enzyme, methylmalonyl-CoA epimerase, which converts the (S) form of methylmalonyl-CoA into the (R) form. This is finally transformed using methylmalonyl-CoA mutase. L-methionine is also metabolized through a longer superpathway (see Figure 2). After transformation to L-homocystein, it is combined with L-serine to make L-cystathione, which is hydrolyzed by cystathione gamma lyase to create 2-oxobutanoate. This substrate is transformed to propanoyl-CoA and undergoes the same metabolism previously described for propanoyl-CoA.[8]
Odd chain fatty acids are also metabolized through the methylmalonyl pathway. The degradation of odd chain fatty acids releases Acetyl-CoA and propionyl-CoA. Propionyl-CoA is then converted to succinyl-CoA, and both succinyl-CoA and propionyl-CoA are interjected into the tricarboxylic acid cycle for continued production of reductant.[9]
Metabolic pathology
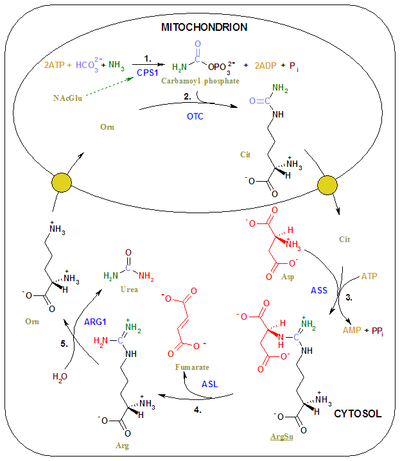
The final product of methylmalonyl-CoA mutase activity is succinyl-CoA which is a tricarboxylic acid cycle substrate. A side effect of excess methylmalonyl-CoA is an interruption of the enzymes responsible for other transformations earlier in the metabolism of propionyl-CoA, leading to propanoic acidemia as well. This, along with an excess of methylmalonyl-CoA leads to the depletion of ATP through oxidative stress and coenzyme A. This disrupts the biosynthesis of myelin, urea, and glucose.[10] Specifically, excess methylmalonyl-CoA places oxidative stress on the mitochondrial enzymes involved in the urea cycle (such as ammonia-dependent-carbamoyl-phosphate synthase or CPS1), and inhibits its mechanism of action.[11] The combination of inhibited urea synthesis and poor protein metabolism, as well as a weakly replenished tricarboxylic acid cycle contribute to the symptoms of methylmalonic acidemia.
Diagnosis and treatment
Several tests can be done to discover the dysfunction of methylmalonyl-CoA mutase. Ammonia test, blood count, CT scan, MRI scan, electrolyte levels, genetic testing, methylmalonic acid blood test, and blood plasma amino acid tests are all can all be conducted to determine deficiency.
There is no treatment for complete lesion of the mut0 gene, though several treatments can help those with slight genetic dysfunction. Liver and kidney transplants, and a low-protein diet all help regulate the effects of the diseases.[12]
Prognosis
Infant mortality is high for patients diagnosed with early onset; mortality can occur within less than 2 months, while children diagnosed with late-onset syndrome seem to have higher rates of survival.[13] Patients suffering from a complete lesion of mut0 have not only the poorest outcome of those suffering from methylaonyl-CoA mutase deficiency, but also of all individuals suffering from any form of methylmalonic acidemia.
See also
External links
- Methylmalonyl-CoA mutase deficiency at NLM Genetics Home Reference
References
- ↑ Online 'Mendelian Inheritance in Man' (OMIM) 251000
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Online 'Mendelian Inheritance in Man' (OMIM) 251100
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 7.0 7.1 7.2 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ 8.0 8.1 Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.
- ↑ Lua error in package.lua at line 80: module 'strict' not found.[dead link]
- ↑ Lua error in package.lua at line 80: module 'strict' not found.